solo per uso di ricerca
Pioglitazone (U-72107) Agonista PPARγ
N. Cat.S2590
Struttura chimica
Peso molecolare: 356.44
Controllo Qualità
| Target correlati | Dehydrogenase HSP Transferase P450 (e.g. CYP17) PDE phosphatase Vitamin Carbohydrate Metabolism Mitochondrial Metabolism Drug Metabolite |
|---|---|
| Altro PPAR Inibitori | T0070907 GW9662 GW6471 WY-14643 (Pirinixic Acid) GSK3787 GW0742 AZ6102 Harmine Astaxanthin Eupatilin |
Coltura cellulare, trattamento e concentrazione di lavoro
| Linee cellulari | Tipo di saggio | Concentrazione | Tempo di incubazione | Formulazione | Descrizione dellattività | PMID |
|---|---|---|---|---|---|---|
| CV-1 | Function assay | In vitro transcriptional activation of Peroxisome proliferator activated receptor gamma (PPAR) expressed in CV-1 cells, EC50=0.69μM | 8576907 | |||
| 3T3-L1 | Function assay | Effective concentration for 50% enhancement of insulin-induced triglyceride accumulation in 3T3-L1 cells, EC50=0.16μM | 9599241 | |||
| CV-1 | Function assay | Activation of peroxisome proliferator activated receptor gamma measured by induction of 50% of maximum alkaline phosphatase activity, transfection assay in CV-1 cells, EC50=0.58884μM | 9836620 | |||
| CV-1 | Function assay | Maximal reporter activity against human Peroxisome proliferator activated receptor gamma Gal4 chimeric in transiently transfected CV-1 cells by functional assay., EC50=0.58μM | 11720854 | |||
| HEK293 | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as induction of receptor interaction with steroid receptor coactivator-1 by EYFP based reporter gene assay | 16680159 | |||
| HEK293 | Function assay | Agonist activity at PPARgamma expressed in HEK293 cells assessed as induction of receptor interaction with retinoid X-receptor alpha by EYFP based reporter gene assay | 16680159 | |||
| CV1 | Function assay | Transactivation of PPARgamma in CV1 cells, EC50=0.55μM | 16821769 | |||
| Cos7 | Function assay | 14 hrs | Transactivation of human PPARgamma LBD expressed in african green monkey Cos7 cells co-transfected with fused GAL4-DBD after 14 hrs by Dual-Glo Luciferase reporter gene assay, EC50=0.3μM | 20307981 | ||
| 3T3L1 | Function assay | 100 umol/L | Increase in PPARgamma mRNA levels in mouse 3T3L1 cells at 100 umol/L by RT-PCR | 20392645 | ||
| CHO | Function assay | Activation of Gal4-tagged human PPARgamma expressed in CHO cells by luciferase reporter gene assay, EC50=0.14μM | 20656494 | |||
| HEK293T | Function assay | 1 uM | 24 hrs | Partial agonist activity at human PPARgamma-LBD expressed in HEK293T cells assessed as induction of receptor transactivation at 1 uM after 24 hrs by luciferase reporter gene assay | 21030263 | |
| COS-1 | Function assay | Agonist activity at human PPARgamma ligand binding domain expressed in COS-1 cells co-transfected with Gal4 by luciferase reporter gene assay, EC50=0.39μM | 21130649 | |||
| CHO | Function assay | 24 hrs | Partial agonist activity at human PPARgamma expressed in CHO cells co-transfected with Gal4-responsive luciferase reporter plasmid after 24 hrs by transactivation assay, EC50=0.39μM | 21377875 | ||
| COS7 | Function assay | Modulation of human PPARgamma-LBD expressed in african green monkey COS7 cells co-transfected with Gal4 assessed as activation of transactivation activity by luciferase assay, EC50=0.3μM | 21873070 | |||
| Sf21 | Function assay | Inhibition of human BSEP expressed in plasma membrane vesicles of Sf21 cells assessed as inhibition of ATP-dependent [3H]taurocholate uptake, IC50=0.3μM | 21965623 | |||
| Sf21 | Function assay | Inhibition of Sprague-Dawley rat Bsep expressed in plasma membrane vesicles of Sf21 cells assessed as inhibition of ATP-dependent [3H]taurocholate uptake, IC50=2μM | 21965623 | |||
| HepG2 | Function assay | 20 hrs | Agonist activity at GAL4-tagged human PPARgamma ligand binding domain expressed in HepG2 cells assessed as transactivation after 20 hrs by beta-galactosidase reporter gene assay, EC50=0.57μM | 22341573 | ||
| HEK293 | Function assay | 18 hrs | Transactivation of human GAL4-fused PPARgamma ligand binding domain transfected in HEK293 cells after 18 hrs by dual luciferase reporter gene assay, EC50=0.8μM | 23102891 | ||
| COS7 | Function assay | Transactivation of GAL4-fused human PPARgamma ligand binding domain transfected in african green monkey COS7 cells by luciferase reporter gene assay, EC50=0.2μM | 23130964 | |||
| THP1 | Antiinflammatory assay | 1 hr | Antiinflammatory activity in human THP1 cells assessed as inhibition of PMA-induced MCP-1 secretion preincubated for 1 hr prior PMA-challenge measured after 48 hrs by ELISA, IC50=18.84μM | 23811092 | ||
| THP1 | Antiinflammatory assay | 2 hrs | Antiinflammatory activity in human THP1 cells assessed as inhibition of PMA-induced MCP-1 secretion incubated for 2 hrs prior to PMA challenge measured after 72 hrs by ELISA, IC50=18.6μM | 24531227 | ||
| HEK293 | Function assay | 10 uM | 24 hrs | Transactivation of PPAR-gamma (unknown origin) expressed in HEK293 cells at 10 uM after 24 hrs by luciferase reporter gene assay | 24890090 | |
| HEK293 | Function assay | 18 hrs | Agonist activity at human PPARgamma expressed in HEK293 cells incubated for 18 hrs by luciferase reporter gene assay, EC50=0.21μM | 25333853 | ||
| COS7 | Function assay | Transactivation of human PPARgamma expressed in African green monkey COS7 cells incubated overnight by dual-glo luciferase reporter gene assay, EC50=0.2μM | 26595749 | |||
| THP1 | Function assay | 10 uM | 24 hrs | Upregulation of ABCA1 mRNA expression in human THP1 cells at 10 uM after 24 hrs by qPCR method relative to control | 27220065 | |
| THP1 | Function assay | 10 uM | 24 hrs | Upregulation of ABCG1 mRNA expression in human THP1 cells at 10 uM after 24 hrs by qPCR method relative to control | 27220065 | |
| COS7 | Function assay | 42 hrs | Transactivation of GAL4-fused human PPARgamma ligand binding domain expressed in African green monkey COS7 cells after 42 hrs by dual luciferase reporter gene assay, EC50=0.5μM | 27560282 | ||
| COS7 | Function assay | 42 hrs | Transactivation at Gal4 fused PPARgamma LBD (unknown origin) expressed in African green monkey COS7 cells after 42 hrs by luciferase assay, EC50=0.32μM | 27569195 | ||
| COS7 | Function assay | 42 hrs | Transactivation of Gal4 fused human PPARgamma LBD expressed in African green monkey COS7 cells after 42 hrs by dual luciferase reporter gene assay, EC50=0.38μM | 27918994 | ||
| PC3 | Function assay | 50 uM | 24 hrs | Increase in p21(WAF1) protein expression in human PC3 cells at 50 uM incubated for 24 hrs by Western blot analysis | 28395220 | |
| MDA-MB-231 | Function assay | 50 uM | 24 hrs | Increase in p21(WAF1) protein expression in human MDA-MB-231 cells at 50 uM incubated for 24 hrs by Western blot analysis | 28395220 | |
| HEK293 | Function assay | 24 hrs | Transactivation activity at Gal4 fused full length human PPARgamma LBD expressed in HEK293 cells after 24 hrs by luciferase reporter gene assay, EC50=1.1μM | 28465099 | ||
| HEK293 | Function assay | 4 hrs | Transrepression activity at human PPARgamma expressed in HEK293 cells assessed as inhibition of TNFalpha induced NF-kappaB promoter activity pretreated for 4 hrs followed by TNFalpha stimulation after 3 hrs by luciferase reporter gene assay, IC50=22μM | 28465099 | ||
| SCC15 | Cytotoxicity assay | 50 uM | 24 hrs | Cytotoxicity against human SCC15 cells transfected with PPARalpha siRNA assessed as reduction in cell viability at 50 uM after 24 hrs by resazurin reduction assay | 29031063 | |
| SCC15 | Cytotoxicity assay | 50 uM | 24 hrs | Cytotoxicity against human SCC15 cells transfected with PPARbeta siRNA assessed as reduction in cell viability at 50 uM after 24 hrs by resazurin reduction assay | 29031063 | |
| HepG2 | Function assay | 24 hrs | Agonist activity at PPARgamma in human HepG2 cells assessed as activation of PPRE incubated for 24 hrs by dual luciferase reporter gene assay, EC50=0.24μM | 30001846 | ||
| HEK293BENA | Function assay | 24 hrs | Transactivation of GAL4-fused human PPARgamma transfected in HEK293BENA cells after 24 hrs by steady glo-luciferase reporter gene assay, EC50=2.053μM | 30351933 | ||
| HEK293 | Function assay | 18 hrs | Transactivation of human full length PPARgamma expressed in HEK293 cells after 18 hrs by luciferase reporter gene based luminescence assay, EC50=0.1μM | 30362739 | ||
| 293H | Function assay | 16 hrs | Transactivation of GAL4-DBD fused human PPARgamma ligand binding domain expressed in UAS-bla HEL 293H cells preincubated for 16 hrs followed by FRET substrate addition and measured after 2 hrs by TR-FRET assay, EC50=0.098μM | 30429097 | ||
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in AP1 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in CD36 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in Glut4 mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| 3T3L1 | Function assay | 10 uM | 24 hrs | Agonist activity at PPARgamma in mouse 3T3L1 cells assessed as increase in adiponectin mRNA expression at 10 uM after 24 hrs by SYBR green dye based RT-PCR analysis | 30594432 | |
| stem cells | Function assay | 5 days | Induction of adipogenesis in human bone marrow-derived mesenchymal stem cells assessed as increase in adiponectin production measured on day 5 in presence of IDX by ELISA, EC50=0.35μM | 30672698 | ||
| HEK293T | Function assay | 18 hrs | Transactivation of full length human PPARgamma2 expressed in HEK293T cells co-expressing PPRE after 18 hrs by luciferase reporter gene assay, EC50=0.25μM | 30676741 | ||
| HEK293T | Function assay | 18 hrs | Transactivation of chimeric Gal4 yeast DBD fused-PPARgamma LBD (unknown origin) expressed in HEK293T cells co-expressing PPRE after 18 hrs by luciferase reporter gene assay, EC50=0.35μM | 30676741 | ||
| HepG2 | Function assay | 30 uM | Binding affinity to NAF1 (unknown origin) expressed in human HepG2 cells assessed as inhibition of mitochondrial respiration at 30 uM | 30770154 | ||
| HepG2 | Function assay | 30 uM | Binding affinity to NAF1 in human HepG2 cells assessed as inhibition of mitochondrial respiration at 30 uM | 30770154 | ||
| COS7 | Function assay | 39 hrs | Transactivation of Gal4-fused human PPARgamma transfected in COS7 cells co-transfected with pGAL5-TK-pGL3 and pRennilla-CMV incubated for 39 hrs by dual luciferase reporter assay, EC50=1.4μM | 31648125 | ||
| K562-IMA[r] | Function assay | 10 uM | 72 hrs | Induction of sensitization to imatinib-induced cytotoxicity in imatinib-resistant human K562-IMA[r] cells assessed as induction of cell death at 10 uM after 72 hrs in presence of 1 uM imatinib by propidium iodide/Triton-X100 dye based FACS analysis | 31648125 | |
| Clicca per visualizzare più dati sperimentali sulle linee cellulari | ||||||
Informazioni chimiche, conservazione e stabilità
| Peso molecolare | 356.44 | Formula | C19H20N2O3S |
Conservazione (Dalla data di ricezione) | |
|---|---|---|---|---|---|
| N. CAS | 111025-46-8 | Scarica SDF | Conservazione delle soluzioni stock |
|
|
| Sinonimi | AD-4833, U 72107 | Smiles | CCC1=CN=C(C=C1)CCOC2=CC=C(C=C2)CC3C(=O)NC(=O)S3 | ||
Solubilità
|
In vitro |
DMSO
: 20 mg/mL
(56.11 mM)
Water : Insoluble Ethanol : Insoluble |
Calcolatore di Molarità
|
In vivo |
|||||
Calcolatore di formulazione in vivo (Soluzione chiara)
Passo 1: Inserire le informazioni di seguito (Consigliato: Un animale aggiuntivo per tenere conto della perdita durante lesperimento)
Passo 2: Inserire la formulazione in vivo (Questo è solo il calcolatore, non la formulazione. Contattateci prima se non cè una formulazione in vivo nella sezione Solubilità.)
Risultati del calcolo:
Concentrazione di lavoro: mg/ml;
Metodo per preparare il liquido master di DMSO: mg farmaco predissolto in μL DMSO ( Concentrazione del liquido master mg/mL, Vi preghiamo di contattarci prima se la concentrazione supera la solubilità del DMSO del lotto del farmaco. )
Metodo per preparare la formulazione in vivo: Prendere μL DMSO liquido master, quindi aggiungereμL PEG300, mescolare e chiarire, quindi aggiungereμL Tween 80, mescolare e chiarire, quindi aggiungere μL ddH2O, mescolare e chiarire.
Metodo per preparare la formulazione in vivo: Prendere μL DMSO liquido master, quindi aggiungere μL Olio di mais, mescolare e chiarire.
Nota: 1. Si prega di assicurarsi che il liquido sia limpido prima di aggiungere il solvente successivo.
2. Assicurarsi di aggiungere il/i solvente/i in ordine. È necessario assicurarsi che la soluzione ottenuta, nellaggiunta precedente, sia una soluzione limpida prima di procedere allaggiunta del solvente successivo. Metodi fisici come il vortex, gli ultrasuoni o il bagno dacqua calda possono essere utilizzati per facilitare la dissoluzione.
Meccanismo dazione
| Targets/IC50/Ki |
PPARγ
|
|---|---|
| In vitro |
Il Pioglitazone viene metabolizzato principalmente dal CYP2C8 e in misura minore dal CYP3A4 in vitro. |
| In vivo |
Il Pioglitazone attenua significativamente la dilatazione e la disfunzione della cavità ventricolare sinistra (LV) mediante ecocardiografia, così come la pressione telediastolica LV in topi con esteso infarto miocardico anteriore. Questo composto normalizza parzialmente LV dP/dt(max) e dP/dt(min), indici della funzione contrattile LV, che sono significativamente ridotti nei topi con IM. Questo composto determina una ridotta attivazione della microglia, una ridotta induzione di cellule positive all'iNOS e meno cellule positive alla proteina acida fibrillare gliale sia nello striato che nella substantia nigra pars compacta del modello murino MPTP della malattia di Parkinson. Blocca quasi completamente la colorazione dei neuroni TH-positivi per la nitrotirosina, un marcatore di danno cellulare mediato dal NO. Questa sostanza chimica (circa 20 mg/kg/giorno) attenua l'attivazione gliale indotta da MPTP e previene la perdita di cellule dopaminergiche nella substantia nigra pars compacta (SNpc) nel modello murino MPTP della malattia di Parkinson. Ne consegue una riduzione del numero di microglia attivate e astrociti reattivi nell'ippocampo e nella corteccia di topi transgenici APPV717I di 10 mesi. Il trattamento con questo composto riduce l'espressione degli enzimi proinfiammatori cicloossigenasi 2 (COX2) e ossido nitrico sintasi inducibile (iNOS). Diminuisce i livelli di mRNA e proteine della beta-secretase-1 (BACE1), e anche una riduzione del 27% dei livelli del peptide Abeta1-42 solubile. |
Riferimenti |
|
Applicazioni
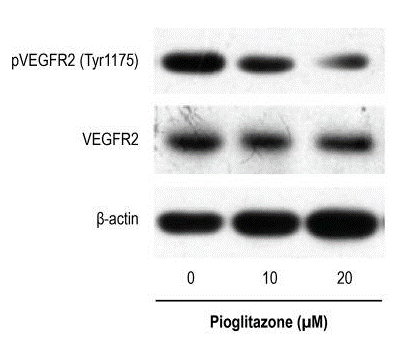
| Metodi | Biomarcatori | Immagini | PMID |
|---|---|---|---|
| Western blot | p-VEGFR2 / VEGFR Phopsho-RB / RB |
|
29972411 |
Informazioni sullo studio clinico
(dati da https://clinicaltrials.gov, aggiornato il 2024-05-22)
| Numero NCT | Reclutamento | Condizioni | Sponsor/Collaboratori | Data di inizio | Fasi |
|---|---|---|---|---|---|
| NCT05946564 | Not yet recruiting | ANCA Associated Vasculitis|Rapidly Progressive Glomerulonephritis|Crescentic Glomerulonephritis |
Assistance Publique - Hôpitaux de Paris|Ministry of Health France |
July 2023 | Phase 3 |
| NCT05305287 | Recruiting | Non-Alcoholic Fatty Liver Disease|Type 2 Diabetes|Mitochondrial Metabolism Disorders |
The University of Texas Health Science Center at San Antonio|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) |
November 1 2022 | Phase 4 |
| NCT05411965 | Completed | Diabetes Mellitus Type 2 |
Boryung Pharmaceutical Co. Ltd |
April 28 2022 | Phase 1 |
| NCT04501406 | Recruiting | Type 2 Diabetes Mellitus (T2DM)|Nonalcoholic Steatohepatitis |
University of Florida|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) |
December 15 2020 | Phase 2 |
| NCT04535700 | Completed | Type 2 Diabetes |
Fundacion para la Investigacion Biomedica del Hospital Universitario Ramon y Cajal |
September 18 2020 | Phase 4 |
| NCT00904046 | Recruiting | Uric Acid Kidney Stone Disease |
University of Texas Southwestern Medical Center|National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)|Takeda Pharmaceuticals North America Inc. |
September 5 2019 | Not Applicable |
Supporto tecnico
Istruzioni per la manipolazione
Tel: +1-832-582-8158 Ext:3
Per qualsiasi altra domanda, si prega di lasciare un messaggio.
